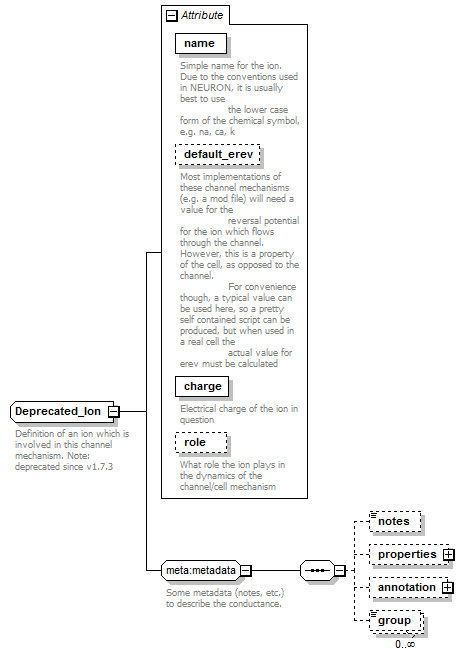
<xs:complexType name="Deprecated_Ion">
<xs:annotation>
<xs:documentation>Definition of an ion which is involved in this channel mechanism. Note: deprecated since v1.7.3</xs:documentation>
</xs:annotation>
<xs:group ref="meta:metadata">
<xs:annotation>
<xs:documentation>Some metadata (notes, etc.) to describe the conductance.</xs:documentation>
</xs:annotation>
</xs:group>
<xs:attribute name="name" type="xs:string" use="required">
<xs:annotation>
<xs:documentation>Simple name for the ion. Due to the conventions used in NEURON, it is usually best to use the lower case form of the chemical symbol, e.g. na, ca, k</xs:documentation>
</xs:annotation>
</xs:attribute>
<xs:attribute name="default_erev" type="bio:VoltageValue" use="optional">
<xs:annotation>
<xs:documentation>Most implementations of these channel mechanisms (e.g. a mod file) will need a value for the reversal potential for the ion which flows through the channel. However, this is a property of the cell, as opposed to the channel. For convenience though, a typical value can be used here, so a pretty self contained script can be produced, but when used in a real cell the actual value for erev must be calculated</xs:documentation>
</xs:annotation>
</xs:attribute>
<xs:attribute name="charge" type="xs:positiveInteger" use="required">
<xs:annotation>
<xs:documentation>Electrical charge of the ion in question</xs:documentation>
</xs:annotation>
</xs:attribute>
<xs:attribute name="role" type="Deprecated_IonRole" use="optional" default="PermeatedSubstance">
<xs:annotation>
<xs:documentation>What role the ion plays in the dynamics of the channel/cell mechanism</xs:documentation>
</xs:annotation>
</xs:attribute>
</xs:complexType> |