Mechanism
Definition of placement of a single electrophysiological mechanism (e.g. channel mechanism) on a group of cables of a cell |
Complex Type Information
Model
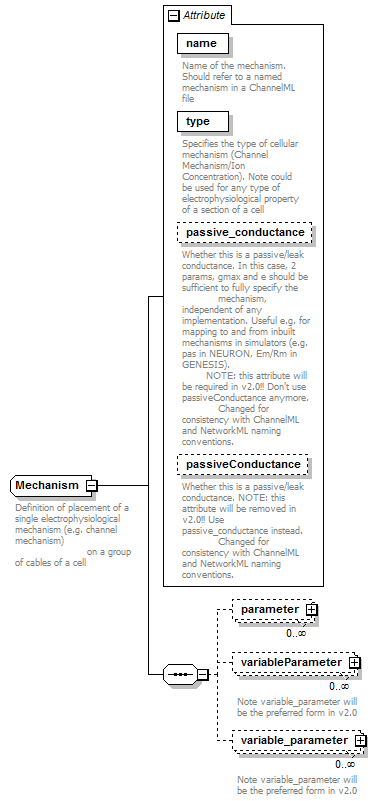
Attributes
| QName | Type | Fixed | Default | Use | Inheritable | Annotation | |
|---|---|---|---|---|---|---|---|
| name | xs:string | required |
|
||||
| passiveConductance | xs:boolean | false | optional |
|
|||
| passive_conductance | xs:boolean | false | optional |
|
|||
| type | MechanismType | required |
|
Used By
| Element | Biophysics/mechanism |
Source
<xs:complexType name="Mechanism"> <xs:annotation> <xs:documentation>Definition of placement of a single electrophysiological mechanism (e.g. channel mechanism) on a group of cables of a cell</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="parameter" type="NamedParameter" minOccurs="0" maxOccurs="unbounded"/> <xs:element name="variableParameter" type="VariableNamedParameter" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Note variable_parameter will be the preferred form in v2.0</xs:documentation> </xs:annotation> </xs:element> <xs:element name="variable_parameter" type="VariableNamedParameter" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Note variable_parameter will be the preferred form in v2.0</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="name" type="xs:string" use="required"> <xs:annotation> <xs:documentation>Name of the mechanism. Should refer to a named mechanism in a ChannelML file</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="type" type="MechanismType" use="required"> <xs:annotation> <xs:documentation>Specifies the type of cellular mechanism (Channel Mechanism/Ion Concentration). Note could be used for any type of electrophysiological property of a section of a cell</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="passive_conductance" type="xs:boolean" use="optional" default="false"> <xs:annotation> <xs:documentation>Whether this is a passive/leak conductance. In this case, 2 params, gmax and e should be sufficient to fully specify the mechanism, independent of any implementation. Useful e.g. for mapping to and from inbuilt mechanisms in simulators (e.g. pas in NEURON, Em/Rm in GENESIS). NOTE: this attribute will be required in v2.0!! Don't use passiveConductance anymore. Changed for consistency with ChannelML and NetworkML naming conventions.</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="passiveConductance" type="xs:boolean" use="optional" default="false"> <xs:annotation> <xs:documentation>Whether this is a passive/leak conductance. NOTE: this attribute will be removed in v2.0!! Use passive_conductance instead. Changed for consistency with ChannelML and NetworkML naming conventions.</xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> |